and the distribution of digital products.
DM Television
Deep Learning Runs on Floating-Point Math. What If That’s a Mistake?
When I first came across the idea of using the Logarithmic Number System (LNS) in deep learning, I was intrigued but also skeptical. Like most of us, I had always worked with Floating-Point (FP) arithmetic—the standard for numerical computation in deep learning. FP provides a good balance between precision and range, but it comes with trade-offs: higher memory usage, increased computational complexity, and greater power consumption. So, I decided to experiment and see for myself—how does LNS compare to FP when training a simple fully connected Multi-Layer Perceptron (MLP) on MNIST?
Why Consider LNS?LNS represents numbers in a logarithmic scale, converting multiplications into additions, which can be computationally cheaper in certain hardware architectures. This efficiency comes at the cost of precision, especially in addition and subtraction operations, which are more complex in LNS. However, the potential benefits—reduced memory footprint, faster computations, and lower power consumption—made me curious enough to try it out.
Background: Floating-Point vs. Logarithmic Number System Floating-Point (FP) RepresentationFloating-Point arithmetic is the standard numerical representation in most deep learning frameworks, such as PyTorch and TensorFlow. FP numbers have:
\
- A sign bit (determining positive or negative value)
- An exponent (scaling factor)
- A mantissa (significand) (precision of the number)
\ FP32 (single precision) is commonly used in deep learning, offering a balance between numerical precision and computational efficiency. More efficient formats like FP16 and BF16 are gaining popularity to accelerate training.
Logarithmic Number System (LNS)LNS is an alternative numerical representation where numbers are stored as logarithms: [ x = \log_b (y) ] where ( b ) is the logarithm base. LNS has several advantages:
\
- Multiplication is simplified to addition: ( x1 * x2 = b^{(\logb x1 + \logb x2)} )
- Division is simplified to subtraction: ( x1 / x2 = b^{(\logb x1 - \logb x2)} )
- Exponential growth functions become linear
\ However, addition and subtraction in LNS require approximations, leading to reduced precision.
LNS Arithmetic OperationsTo further explore LNS, I implemented basic arithmetic operations such as addition, subtraction, multiplication, and division using LNS internal representations.
\
import torch import numpy as np import xlns as xl # Assuming xlns module is installed and provides xlnsnp # Function to convert floating-point numbers to xlns internal representation def float_to_internal(arr): xlns_data = xl.xlnsnp(arr) return xlns_data.nd # Function to convert xlns internal representation back to floating-point numbers def internal_to_float(internal_data): original_numbers = [] for value in internal_data: x = value // 2 s = value % 2 # Use x and s to create xlns object xlns_value = xl.xlns(0) xlns_value.x = x xlns_value.s = s original_numbers.append(float(xlns_value)) return original_numbers # Function to perform LNS addition using internal representation def lns_add_internal(x, y): max_part = torch.maximum(x, y) diff = torch.abs(x - y) adjust_term = torch.log1p(torch.exp(-diff)) return max_part + adjust_term # Function to perform LNS subtraction using internal representation def lns_sub_internal(x, y): return lns_add_internal(x, -y) # Function to perform LNS multiplication using internal representation def lns_mul_internal(x, y): return x + y # Function to perform LNS division using internal representation def lns_div_internal(x, y): return x - y # Input floating-point arrays x_float = [2.0, 3.0] y_float = [-1.0, 0.0] # Convert floating-point arrays to xlns internal representation x_internal = float_to_internal(x_float) y_internal = float_to_internal(y_float) # Create tensors from the internal representation tensor_x_nd = torch.tensor(x_internal, dtype=torch.int64) tensor_y_nd = torch.tensor(y_internal, dtype=torch.int64) # Perform the toy LNS addition on the internal representation result_add_internal = lns_add_internal(tensor_x_nd, tensor_y_nd) # Perform the toy LNS subtraction on the internal representation result_sub_internal = lns_sub_internal(tensor_x_nd, tensor_y_nd) # Perform the toy LNS multiplication on the internal representation result_mul_internal = lns_mul_internal(tensor_x_nd, tensor_y_nd) # Perform the toy LNS division on the internal representation result_div_internal = lns_div_internal(tensor_x_nd, tensor_y_nd) # Convert the internal results back to original floating-point values result_add_float = internal_to_float(result_add_internal.numpy()) result_sub_float = internal_to_float(result_sub_internal.numpy()) result_mul_float = internal_to_float(result_mul_internal.numpy()) result_div_float = internal_to_float(result_div_internal.numpy()) # Convert the results back to PyTorch tensors result_add_tensor = torch.tensor(result_add_float, dtype=torch.float32) result_sub_tensor = torch.tensor(result_sub_float, dtype=torch.float32) result_mul_tensor = torch.tensor(result_mul_float, dtype=torch.float32) result_div_tensor = torch.tensor(result_div_float, dtype=torch.float32) # Print results print("Input x:", x_float) print("Input y:", y_float) print("Addition Result:", result_add_float) print("Addition Result Tensor:", result_add_tensor) print("Subtraction Result:", result_sub_float) print("Subtraction Result Tensor:", result_sub_tensor) print("Multiplication Result:", result_mul_float) print("Multiplication Result Tensor:", result_mul_tensor) print("Division Result:", result_div_float) print("Division Result Tensor:", result_div_tensor)\ Here’s a breakdown of my experimental implementation of the Logarithmic Number System (LNS).
1. Basic LNS Concept and Challenges in PyTorchIn LNS, numbers are represented as logarithms, which transforms multiplication and division into addition and subtraction. However, implementing this with PyTorch presents specific challenges since PyTorch tensors use floating-point representations internally. This creates several requirements:
\
- Maintain logarithmic representation throughout computations.
- Ensure numerical stability.
- Handle conversions carefully.
- Manage the internal representation using two components:
- x: the logarithmic value.
- s: a sign bit (0 or 1).
The first step is converting floating-point numbers to their LNS internal representation.
import torch import numpy as np import xl # Hypothetical external LNS library def float_to_internal(arr): xlns_data = xl.xlnsnp(arr) return xlns_data.nd # Convert floating-point arrays to xlns internal representation x_float = np.array([2.0, 3.0]) y_float = np.array([-1.0, 0.0]) x_internal = float_to_internal(x_float) y_internal = float_to_internal(y_float) # Create tensors from the internal representation tensor_x_nd = torch.tensor(x_internal, dtype=torch.int64) tensor_y_nd = torch.tensor(y_internal, dtype=torch.int64)\ The use of dtype=torch.int64 is crucial because:
- It preserves the exact LNS internal representation without floating-point rounding errors.
- Packs both logarithmic value and sign bit into a single integer.
- Prevents unintended floating-point operations from corrupting the LNS representation.
Multiplication in LNS becomes addition:
- If a = log(x) and b = log(y), then log(x×y) = log(x) + log(y).
Division becomes subtraction:
- log(x/y) = log(x) - log(y).
\ Addition is more complex and numerically sensitive because:
- It involves exponential and logarithmic operations.
- Direct floating-point implementation could lead to overflow/underflow.
- Uses the equation: log(x + y) = log(max(x,y)) + log(1 + exp(log(min(x,y)) - log(max(x,y)))).
- Uses log1p instead of direct log(1 + x) for better numerical stability.
\ The conversion pipeline maintains a clear separation:
- Convert from float → LNS internal representation (integers).
- Perform LNS operations using integer arithmetic.
- Convert back to float only when necessary.
- Multiplication and division are simplified to addition and subtraction.
- Wide dynamic range with fixed-point arithmetic.
- Potentially more efficient for certain applications.
- Addition and subtraction are more complex operations.
- Conversion overhead between floating-point and LNS.
- Requires special handling for zero and negative numbers.
- PyTorch tensor compatibility requires careful type management.
To improve performance, one could:
- Implement a custom PyTorch autograd function for LNS operations.
- Create a custom tensor type that natively supports LNS.
- Use CUDA kernels for efficient LNS operations on GPU.
\ The current implementation makes practical trade-offs:
- Prioritizes clarity and maintainability over maximum performance.
- Uses PyTorch's existing tensor infrastructure while preserving LNS accuracy.
- Maintains numerical stability through careful type management.
- Minimizes conversions between representations.
The following steps demonstrate the complete pipeline using example values [2.0, 3.0] and [-1.0, 0.0]:
- Convert input floats to LNS internal representation.
- Create integer tensors to store LNS values.
- Perform arithmetic operations in LNS domain.
- Convert results back to floating-point.
- Create final PyTorch tensors for further processing.
\ This implementation successfully bridges the gap between PyTorch's floating-point tensor system and LNS arithmetic while maintaining numerical stability and accuracy.
Training a fully connected MLP on the MNIST Digit dataset with FP and LNS Experiment SetupI trained a fully connected MLP on the MNIST dataset using both FP and LNS representations. The model architecture was simple:
- Input layer: 784 neurons (flattened 28x28 images)
- Hidden layers: Two layers with 256 and 128 neurons, ReLU activations
- Output layer: 10 neurons (one for each digit, using softmax)
- Loss function: Cross-entropy
- Optimizer: Adam
\ For the LNS implementation, I had to step outside of my usual PyTorch workflow. Unlike FP, which PyTorch natively supports, PyTorch does not provide built-in LNS operations. I found a GitHub project called xlns, which implements logarithmic number representations and arithmetic, making it possible to use LNS in neural networks.
Floating-Point MLP in PyTorchWe start by implementing a standard FP-based fully connected MLP using PyTorch:
import torch import torch.nn as nn import torch.optim as optim import torchvision import torchvision.transforms as transforms import matplotlib.pyplot as plt import numpy as np import time # For calculating elapsed time # Define the multi-layer perceptron (MLP) model with one hidden layer class MLP(nn.Module): def __init__(self): super(MLP, self).__init__() # Input: 28*28 features; Hidden layer: 100 neurons; Output layer: 10 neurons self.fc1 = nn.Linear(28 * 28, 100) self.relu = nn.ReLU() self.fc2 = nn.Linear(100, 10) self.logsoftmax = nn.LogSoftmax(dim=1) # For stable outputs with NLLLoss def forward(self, x): # Flatten image: (batch_size, 1, 28, 28) -> (batch_size, 784) x = x.view(x.size(0), -1) x = self.fc1(x) x = self.relu(x) x = self.fc2(x) return self.logsoftmax(x) def train_and_validate(num_epochs=5, batch_size=64, learning_rate=0.01, split_ratio=0.8): # Set the device to GPU if available device = torch.device("cuda" if torch.cuda.is_available() else "cpu") print(f"Training on device: {device}") # Transformation for MNIST: convert to tensor and normalize transform = transforms.Compose([ transforms.ToTensor(), transforms.Normalize((0.5,), (0.5,)) ]) # Load the MNIST training dataset train_dataset_full = torchvision.datasets.MNIST( root='./data', train=True, transform=transform, download=True ) # Split the dataset into training and validation sets n_total = len(train_dataset_full) n_train = int(split_ratio * n_total) n_val = n_total - n_train train_dataset, val_dataset = torch.utils.data.random_split(train_dataset_full, [n_train, n_val]) # Create DataLoaders for training and validation datasets train_loader = torch.utils.data.DataLoader(train_dataset, batch_size=batch_size, shuffle=True) val_loader = torch.utils.data.DataLoader(val_dataset, batch_size=batch_size, shuffle=False) # Initialize the model, loss function, and optimizer; move model to device model = MLP().to(device) criterion = nn.NLLLoss() optimizer = optim.SGD(model.parameters(), lr=learning_rate) # Lists to store training and validation accuracies for each epoch train_accuracies = [] val_accuracies = [] # Record the start time for measuring elapsed time start_time = time.time() # Training loop for epoch in range(num_epochs): model.train() running_loss = 0.0 correct_train = 0 total_train = 0 for inputs, labels in train_loader: # Move inputs and labels to device inputs, labels = inputs.to(device), labels.to(device) optimizer.zero_grad() outputs = model(inputs) loss = criterion(outputs, labels) loss.backward() optimizer.step() # Compute running loss and training accuracy running_loss += loss.item() * inputs.size(0) _, predicted = torch.max(outputs.data, 1) total_train += labels.size(0) correct_train += (predicted == labels).sum().item() train_accuracy = 100.0 * correct_train / total_train train_accuracies.append(train_accuracy) # Evaluate on validation set model.eval() correct_val = 0 total_val = 0 with torch.no_grad(): for inputs, labels in val_loader: inputs, labels = inputs.to(device), labels.to(device) outputs = model(inputs) _, predicted = torch.max(outputs.data, 1) total_val += labels.size(0) correct_val += (predicted == labels).sum().item() val_accuracy = 100.0 * correct_val / total_val val_accuracies.append(val_accuracy) print(f"Epoch [{epoch+1}/{num_epochs}], Loss: {running_loss/total_train:.4f}, " f"Train Acc: {train_accuracy:.2f}%, Val Acc: {val_accuracy:.2f}%") # Calculate elapsed time elapsed_time = time.time() - start_time print(f"Training completed in {elapsed_time:.2f} seconds.") # Show sample predictions from the validation set show_predictions(model, val_loader, device) # Optional: plot training and validation accuracies epochs_arr = np.arange(1, num_epochs + 1) plt.figure(figsize=(10, 6)) plt.plot(epochs_arr, train_accuracies, label='Training Accuracy', marker='o') plt.plot(epochs_arr, val_accuracies, label='Validation Accuracy', marker='x') plt.xlabel('Epoch') plt.ylabel('Accuracy (%)') plt.title('Training and Validation Accuracies') plt.legend() plt.grid(True) plt.savefig('pytorch_accuracy.png') plt.show() def show_predictions(model, data_loader, device, num_images=6): """ Displays a few sample images from the data_loader along with the model's predictions. """ model.eval() images_shown = 0 plt.figure(figsize=(12, 8)) # Get one batch of images from the validation dataset for inputs, labels in data_loader: inputs, labels = inputs.to(device), labels.to(device) with torch.no_grad(): outputs = model(inputs) _, predicted = torch.max(outputs, 1) # Loop through the batch and plot images for i in range(inputs.size(0)): if images_shown >= num_images: break # Move the image to cpu and convert to numpy for plotting img = inputs[i].cpu().squeeze() plt.subplot(2, num_images // 2, images_shown + 1) plt.imshow(img, cmap='gray') plt.title(f"Pred: {predicted[i].item()}") plt.axis('off') images_shown += 1 if images_shown >= num_images: break plt.suptitle("Sample Predictions from the Validation Set") plt.tight_layout() plt.show() if __name__ == '__main__': train_and_validate(num_epochs=5, batch_size=64, learning_rate=0.01, split_ratio=0.8)\ This implementation follows a conventional deep learning pipeline where multiplications and additions are handled by FP arithmetic.
\ Here is a detailed walkthrough of this PyTorch implementation of a Multi-Layer Perceptron (MLP) for the MNIST dataset.
- Model Architecture (MLP Class):
- Forward Pass:
- Training Setup:
Key components:
GPU support through device selection
Data normalization for better training
Configurable hyperparameters
\
- Dataset Management:
Downloads MNIST dataset if not present
Splits data into training (80%) and validation (20%) sets
\
- Training Loop:
Classic training procedure:
Zero gradients
Forward pass
Loss computation
Backward pass
Weight updates
\
- Validation:
Key features:
Model set to evaluation mode
No gradient computation needed
Accuracy calculation
\
- Visualization:
Shows sample predictions from validation set
Useful for qualitative assessment
\
- Performance Tracking:
Tracks training and validation accuracies
Plots learning curves
Measures training time
\
This provides a solid foundation for comparing with LNS-based implementations, as it implements all standard components of a deep learning pipeline using traditional floating-point arithmetic.
Logarithmic Number System (LNS) MLPFor LNS, we need to use the xlns library. Unlike FP, LNS replaces multiplication-heavy operations with addition in the logarithmic domain. However, PyTorch does not natively support this, so we have to manually apply LNS operations where applicable.
import numpy as np import matplotlib.pyplot as plt import os import time import argparse import xlns as xl from tensorflow.keras.datasets import mnist # Use Keras's MNIST loader # If you are using fractional normalized LNS, make sure the following are uncommented import xlnsconf.xlnsudFracnorm xlnsconf.xlnsudFracnorm.ilog2 = xlnsconf.xlnsudFracnorm.ipallog2 xlnsconf.xlnsudFracnorm.ipow2 = xlnsconf.xlnsudFracnorm.ipalpow2 # Set global parameter in xlns xl.xlnssetF(10) def softmax(inp): max_vals = inp.max(axis=1) max_vals = xl.reshape(max_vals, (xl.size(max_vals), 1)) u = xl.exp(inp - max_vals) v = u.sum(axis=1) v = v.reshape((xl.size(v), 1)) u = u / v return u def main(main_params): print("arbitrary base np LNS. Also xl.hstack, xl routines in softmax") print("testing new softmax and * instead of @ for delta") print("works with type " + main_params['type']) is_training = bool(main_params['is_training']) leaking_coeff = float(main_params['leaking_coeff']) batchsize = int(main_params['minibatch_size']) lr = float(main_params['learning_rate']) num_epoch = int(main_params['num_epoch']) _lambda = float(main_params['lambda']) ones = np.array(list(np.ones((batchsize, 1)))) if is_training: # Load the MNIST dataset from Keras (x_train, y_train), (x_test, y_test) = mnist.load_data() # Normalize images to [0, 1] x_train = x_train.astype(np.float64) / 255.0 x_test = x_test.astype(np.float64) / 255.0 # One-hot encode the labels (assume 10 classes for MNIST digits 0-9) num_classes = 10 y_train = np.eye(num_classes)[y_train] y_test = np.eye(num_classes)[y_test] # Flatten the images from (28, 28) to (784,) x_train = x_train.reshape(x_train.shape[0], -1) x_test = x_test.reshape(x_test.shape[0], -1) # Use a portion of the training data for validation (the 'split' index) split = int(main_params['split']) x_val = x_train[split:] y_val = y_train[split:] x_train = x_train[:split] y_train = y_train[:split] # If available, load pretrained weights; otherwise, initialize new random weights. if os.path.isfile("./weightin.npz"): print("using ./weightin.npz") randfile = np.load("./weightin.npz", "r") W1 = randfile["W1"] W2 = randfile["W2"] randfile.close() else: print("using new random weights") # Note: The input layer now has 785 neurons (784 features + 1 bias). W1 = np.array(list(np.random.normal(0, 0.1, (785, 100)))) # The first hidden layer has 100 neurons; add bias so 101 W2 = np.array(list(np.random.normal(0, 0.1, (101, 10)))) np.savez_compressed("./weightout.npz", W1=W1, W2=W2) delta_W1 = np.array(list(np.zeros(W1.shape))) delta_W2 = np.array(list(np.zeros(W2.shape))) # Convert weights to desired type (xlns variants or float) if main_params['type'] == 'xlnsnp': lnsW1 = xl.xlnsnp(np.array(xl.xlnscopy(list(W1)))) lnsW2 = xl.xlnsnp(np.array(xl.xlnscopy(list(W2)))) lnsones = xl.xlnsnp(np.array(xl.xlnscopy(list(np.ones((batchsize, 1)))))) lnsdelta_W1 = xl.xlnsnp(np.array(xl.xlnscopy(list(np.zeros(W1.shape))))) lnsdelta_W2 = xl.xlnsnp(np.array(xl.xlnscopy(list(np.zeros(W2.shape))))) elif main_params['type'] == 'xlnsnpv': lnsW1 = xl.xlnsnpv(np.array(xl.xlnscopy(list(W1))), 6) lnsW2 = xl.xlnsnpv(np.array(xl.xlnscopy(list(W2))), 6) lnsones = xl.xlnsnpv(np.array(xl.xlnscopy(list(np.ones((batchsize, 1)))))) lnsdelta_W1 = xl.xlnsnpv(np.array(xl.xlnscopy(list(np.zeros(W1.shape))))) lnsdelta_W2 = xl.xlnsnpv(np.array(xl.xlnscopy(list(np.zeros(W2.shape))))) elif main_params['type'] == 'xlnsnpb': lnsW1 = xl.xlnsnpb(np.array(xl.xlnscopy(list(W1))), 2**2**-6) lnsW2 = xl.xlnsnpb(np.array(xl.xlnscopy(list(W2))), 2**2**-6) lnsones = xl.xlnsnpb(np.array(xl.xlnscopy(list(np.ones((batchsize, 1))))), 2**2**-xl.xlnsF) lnsdelta_W1 = xl.xlnsnpb(np.array(xl.xlnscopy(list(np.zeros(W1.shape)))), 2**2**-xl.xlnsF) lnsdelta_W2 = xl.xlnsnpb(np.array(xl.xlnscopy(list(np.zeros(W2.shape)))), 2**2**-xl.xlnsF) elif main_params['type'] == 'xlns': lnsW1 = np.array(xl.xlnscopy(list(W1))) lnsW2 = np.array(xl.xlnscopy(list(W2))) lnsones = np.array(xl.xlnscopy(list(np.ones((batchsize, 1))))) lnsdelta_W1 = np.array(xl.xlnscopy(list(np.zeros(W1.shape)))) lnsdelta_W2 = np.array(xl.xlnscopy(list(np.zeros(W2.shape)))) elif main_params['type'] == 'xlnsud': lnsW1 = np.array(xl.xlnscopy(list(W1), xl.xlnsud)) lnsW2 = np.array(xl.xlnscopy(list(W2), xl.xlnsud)) lnsones = np.array(xl.xlnscopy(list(np.ones((batchsize, 1))), xl.xlnsud)) lnsdelta_W1 = np.array(xl.xlnscopy(list(np.zeros(W1.shape)), xl.xlnsud)) lnsdelta_W2 = np.array(xl.xlnscopy(list(np.zeros(W2.shape)), xl.xlnsud)) elif main_params['type'] == 'xlnsv': lnsW1 = np.array(xl.xlnscopy(list(W1), xl.xlnsv, 6)) lnsW2 = np.array(xl.xlnscopy(list(W2), xl.xlnsv, 6)) lnsones = np.array(xl.xlnscopy(list(np.ones((batchsize, 1))), xl.xlnsv)) lnsdelta_W1 = np.array(xl.xlnscopy(list(np.zeros(W1.shape)), xl.xlnsv)) lnsdelta_W2 = np.array(xl.xlnscopy(list(np.zeros(W2.shape)), xl.xlnsv)) elif main_params['type'] == 'xlnsb': lnsW1 = np.array(xl.xlnscopy(list(W1), xl.xlnsb, 2**2**-6)) lnsW2 = np.array(xl.xlnscopy(list(W2), xl.xlnsb, 2**2**-6)) lnsones = np.array(xl.xlnscopy(list(np.ones((batchsize, 1))), xl.xlnsb, 2**2**-xl.xlnsF)) lnsdelta_W1 = np.array(xl.xlnscopy(list(np.zeros(W1.shape)), xl.xlnsb, 2**2**-xl.xlnsF)) lnsdelta_W2 = np.array(xl.xlnscopy(list(np.zeros(W2.shape)), xl.xlnsb, 2**2**-xl.xlnsF)) elif main_params['type'] == 'float': lnsW1 = np.array(list(W1)) lnsW2 = np.array(list(W2)) lnsones = np.array(list(np.ones((batchsize, 1)))) lnsdelta_W1 = np.array(list(np.zeros(W1.shape))) lnsdelta_W2 = np.array(list(np.zeros(W2.shape))) performance = {} performance['lnsacc_train'] = np.zeros(num_epoch) performance['lnsacc_val'] = np.zeros(num_epoch) start_time = time.process_time() # Training loop for epoch in range(num_epoch): print('At Epoch %d:' % (1 + epoch)) # Loop through training batches for mbatch in range(int(split / batchsize)): start = mbatch * batchsize x = np.array(x_train[start:(start + batchsize)]) y = np.array(y_train[start:(start + batchsize)]) # At this point, each x is already flattened (batchsize x 784) # Conversion based on type if main_params['type'] == 'xlnsnp': lnsx = xl.xlnsnp(np.array(xl.xlnscopy(np.array(x, dtype=np.float64)))) lnsy = xl.xlnsnp(np.array(xl.xlnscopy(np.array(y, dtype=np.float64)))) elif main_params['type'] == 'xlnsnpv': lnsx = xl.xlnsnpv(np.array(xl.xlnscopy(np.array(x, dtype=np.float64)))) lnsy = xl.xlnsnpv(np.array(xl.xlnscopy(np.array(y, dtype=np.float64)))) elif main_params['type'] == 'xlnsnpb': lnsx = xl.xlnsnpb(np.array(xl.xlnscopy(np.array(x, dtype=np.float64))), 2**2**-xl.xlnsF) lnsy = xl.xlnsnpb(np.array(xl.xlnscopy(np.array(y, dtype=np.float64))), 2**2**-xl.xlnsF) elif main_params['type'] == 'xlns': lnsx = np.array(xl.xlnscopy(np.array(x, dtype=np.float64))) lnsy = np.array(xl.xlnscopy(np.array(y, dtype=np.float64))) elif main_params['type'] == 'xlnsud': lnsx = np.array(xl.xlnscopy(np.array(x, dtype=np.float64), xl.xlnsud)) lnsy = np.array(xl.xlnscopy(np.array(y, dtype=np.float64), xl.xlnsud)) elif main_params['type'] == 'xlnsv': lnsx = np.array(xl.xlnscopy(np.array(x, dtype=np.float64), xl.xlnsv)) lnsy = np.array(xl.xlnscopy(np.array(y, dtype=np.float64), xl.xlnsv)) elif main_params['type'] == 'xlnsb': lnsx = np.array(xl.xlnscopy(np.array(x, dtype=np.float64), xl.xlnsv, 2**2**-xl.xlnsF)) lnsy = np.array(xl.xlnscopy(np.array(y, dtype=np.float64), xl.xlnsv, 2**2**-xl.xlnsF)) elif main_params['type'] == 'float': lnsx = np.array(x, dtype=np.float64) lnsy = np.array(y, dtype=np.float64) # Concatenate the bias "ones" with input features for the first layer lnss1 = xl.hstack((lnsones, lnsx)) @ lnsW1 lnsmask = (lnss1 > 0) + (leaking_coeff * (lnss1 < 0)) lnsa1 = lnss1 * lnsmask lnss2 = xl.hstack((lnsones, lnsa1)) @ lnsW2 lnsa2 = softmax(lnss2) lnsgrad_s2 = (lnsa2 - lnsy) / batchsize lnsgrad_a1 = lnsgrad_s2 @ xl.transpose(lnsW2[1:]) lnsdelta_W2 = xl.transpose(xl.hstack((lnsones, lnsa1))) * lnsgrad_s2 lnsgrad_s1 = lnsmask * lnsgrad_a1 lnsdelta_W1 = xl.transpose(xl.hstack((lnsones, lnsx))) * lnsgrad_s1 lnsW2 -= (lr * (lnsdelta_W2 + (_lambda * lnsW2))) lnsW1 -= (lr * (lnsdelta_W1 + (_lambda * lnsW1))) print('#= ', split, ' batch=', batchsize, ' lr=', lr) lnscorrect_count = 0 # Evaluate accuracy on training set for mbatch in range(int(split / batchsize)): start = mbatch * batchsize x = x_train[start:(start + batchsize)] y = y_train[start:(start + batchsize)] if main_params['type'] == 'xlnsnp': lnsx = xl.xlnsnp(np.array(xl.xlnscopy(np.array(x, dtype=np.float64)))) elif main_params['type'] == 'xlnsnpv': lnsx = xl.xlnsnpv(np.array(xl.xlnscopy(np.array(x, dtype=np.float64)))) elif main_params['type'] == 'xlnsnpb': lnsx = xl.xlnsnpb(np.array(xl.xlnscopy(np.array(x, dtype=np.float64))), 2**2**-xl.xlnsF) elif main_params['type'] == 'xlns': lnsx = np.array(xl.xlnscopy(np.array(x, dtype=np.float64))) elif main_params['type'] == 'xlnsud': lnsx = np.array(xl.xlnscopy(np.array(x, dtype=np.float64), xl.xlnsud)) elif main_params['type'] == 'xlnsv': lnsx = np.array(xl.xlnscopy(np.array(x, dtype=np.float64), xl.xlnsv)) elif main_params['type'] == 'xlnsb': lnsx = np.array(xl.xlnscopy(np.array(x, dtype=np.float64), xl.xlnsv, 2**2**-xl.xlnsF)) elif main_params['type'] == 'float': lnsx = np.array(x, dtype=np.float64) lnss1 = xl.hstack((lnsones, lnsx)) @ lnsW1 lnsmask = (lnss1 > 0) + (leaking_coeff * (lnss1 < 0)) lnsa1 = lnss1 * lnsmask lnss2 = xl.hstack((lnsones, lnsa1)) @ lnsW2 lnscorrect_count += np.sum(np.argmax(y, axis=1) == xl.argmax(lnss2, axis=1)) lnsaccuracy = lnscorrect_count / split print("train-set accuracy at epoch %d: %f" % (1 + epoch, lnsaccuracy)) performance['lnsacc_train'][epoch] = 100 * lnsaccuracy lnscorrect_count = 0 # Evaluate on the validation set for mbatch in range(int(split / batchsize)): start = mbatch * batchsize x = x_val[start:(start + batchsize)] y = y_val[start:(start + batchsize)] if main_params['type'] == 'xlnsnp': lnsx = xl.xlnsnp(np.array(xl.xlnscopy(np.array(x, dtype=np.float64)))) elif main_params['type'] == 'xlnsnpv': lnsx = xl.xlnsnpv(np.array(xl.xlnscopy(np.array(x, dtype=np.float64)))) elif main_params['type'] == 'xlnsnpb': lnsx = xl.xlnsnpb(np.array(xl.xlnscopy(np.array(x, dtype=np.float64))), 2**2**-xl.xlnsF) elif main_params['type'] == 'xlns': lnsx = np.array(xl.xlnscopy(np.array(x, dtype=np.float64))) elif main_params['type'] == 'xlnsud': lnsx = np.array(xl.xlnscopy(np.array(x, dtype=np.float64), xl.xlnsud)) elif main_params['type'] == 'xlnsv': lnsx = np.array(xl.xlnscopy(np.array(x, dtype=np.float64), xl.xlnsv)) elif main_params['type'] == 'xlnsb': lnsx = np.array(xl.xlnscopy(np.array(x, dtype=np.float64), xl.xlnsv, 2**2**-xl.xlnsF)) elif main_params['type'] == 'float': lnsx = np.array(x, dtype=np.float64) lnss1 = xl.hstack((lnsones, lnsx)) @ lnsW1 lnsmask = (lnss1 > 0) + (leaking_coeff * (lnss1 < 0)) lnsa1 = lnss1 * lnsmask lnss2 = xl.hstack((lnsones, lnsa1)) @ lnsW2 lnscorrect_count += np.sum(np.argmax(y, axis=1) == xl.argmax(lnss2, axis=1)) lnsaccuracy = lnscorrect_count / split print("Val-set accuracy at epoch %d: %f" % (1 + epoch, lnsaccuracy)) performance['lnsacc_val'][epoch] = 100 * lnsaccuracy print("elapsed time=" + str(time.process_time() - start_time)) fig = plt.figure(figsize=(16, 9)) ax = fig.add_subplot(111) x_axis = range(1, 1 + performance['lnsacc_train'].size) ax.plot(x_axis, performance['lnsacc_train'], 'y') ax.plot(x_axis, performance['lnsacc_val'], 'm') ax.set_xlabel('Number of Epochs') ax.set_ylabel('Accuracy') plt.suptitle(main_params['type'] + ' ' + str(split) + ' Validation and Training MNIST Accuracies F=' + str(xl.xlnsF), fontsize=14) ax.legend(['train', 'validation']) plt.grid(which='both', axis='both', linestyle='-.') plt.savefig('genericaccuracy.png') plt.show() # Now, show predictions on a few test images num_examples = 5 # Number of test images to display selected_indices = np.arange(num_examples) # choose the first few images for demo x_sample = x_test[selected_indices] y_sample = y_test[selected_indices] # For prediction, create a bias vector matching the sample size ones_sample = np.ones((x_sample.shape[0], 1)) z1_sample = np.hstack((ones_sample, x_sample)) @ lnsW1 mask_sample = (z1_sample > 0) + (leaking_coeff * (z1_sample < 0)) a1_sample = z1_sample * mask_sample z2_sample = np.hstack((ones_sample, a1_sample)) @ lnsW2 pred_probs = softmax(z2_sample) predictions = np.argmax(pred_probs, axis=1) true_labels = np.argmax(y_sample, axis=1) # Plot each test image along with its prediction and true label plt.figure(figsize=(10, 2)) for i in range(num_examples): plt.subplot(1, num_examples, i + 1) # Reshape the flattened image back to 28x28 for display plt.imshow(x_sample[i].reshape(28, 28), cmap='gray') plt.title(f"Pred: {predictions[i]}\nTrue: {true_labels[i]}") plt.axis('off') plt.tight_layout() plt.show() if __name__ == '__main__': # In a Kaggle notebook, set parameters manually using a dictionary. main_params = { 'is_training': True, 'split': 50, 'learning_rate': 0.01, 'lambda': 0.000, 'minibatch_size': 1, 'num_epoch': 5, 'leaking_coeff': 0.0078125, 'type': 'float' } main(main_params)\ I shall walk you through this code which implements a Logarithmic Number System (LNS) Multi-Layer Perceptron (MLP) for MNIST digit classification. Let me break it down into key sections:
\
- Setup and Imports:
The code uses the xlns library for logarithmic number system operations
It offers multiple LNS variants (xlnsnp, xlnsnpv, xlnsud, etc.) for different precision and performance tradeoffs
The MNIST dataset is loaded through Keras
\
- Core Functions:
This is a numerically stable softmax implementation adapted for LNS operations.
\
- Network Architecture:
Input layer: 784 neurons (28x28 flattened MNIST images) + 1 bias = 785
Hidden layer: 100 neurons + 1 bias = 101
Output layer: 10 neurons (one per digit)
\
- Weight Initialization:
Weights are either loaded from a file ("weightin.npz") or initialized randomly
Random weights use normal distribution with mean=0, std=0.1
Different LNS variants require different initialization methods (xlnsnp, xlnsnpv, etc.)
\
- Training Loop:
\ Key aspects of the training:
Uses leaky ReLU activation (controlled by leaking_coeff)
Implements standard backpropagation but with LNS operations
Includes L2 regularization (lambda parameter)
Updates weights using gradient descent with learning rate 'lr'
\
- Evaluation:
Tracks both training and validation accuracy
Plots learning curves showing accuracy over epochs
Displays sample predictions on test images
\
- Hyperparameters:
Uses mini-batch gradient descent (default batch size = 1)
Implements early stopping through validation set splitting
Leaky ReLU coefficient is set to 0.0078125
\
- Visualization:
- Creates plots showing training and validation accuracy
- Displays sample test images with predictions and true labels
- Saves accuracy plot as 'genericaccuracy.png'
\ The key innovation here is the use of LNS arithmetic which replaces multiplications with additions in the log domain, potentially offering better computational efficiency for certain hardware implementations. The code supports multiple LNS variants allowing for different precision-performance tradeoffs.
Basic Performance Comparison Floating-Point Model Performance Training on device: cuda Epoch [1/5], Loss: 0.8540, Train Acc: 79.60%, Val Acc: 88.22% Epoch [2/5], Loss: 0.3917, Train Acc: 88.97%, Val Acc: 89.92% Epoch [3/5], Loss: 0.3380, Train Acc: 90.29%, Val Acc: 90.60% Epoch [4/5], Loss: 0.3104, Train Acc: 90.96%, Val Acc: 91.12% Epoch [5/5], Loss: 0.2901, Train Acc: 91.60%, Val Acc: 91.62% Training completed in 57.76 seconds.
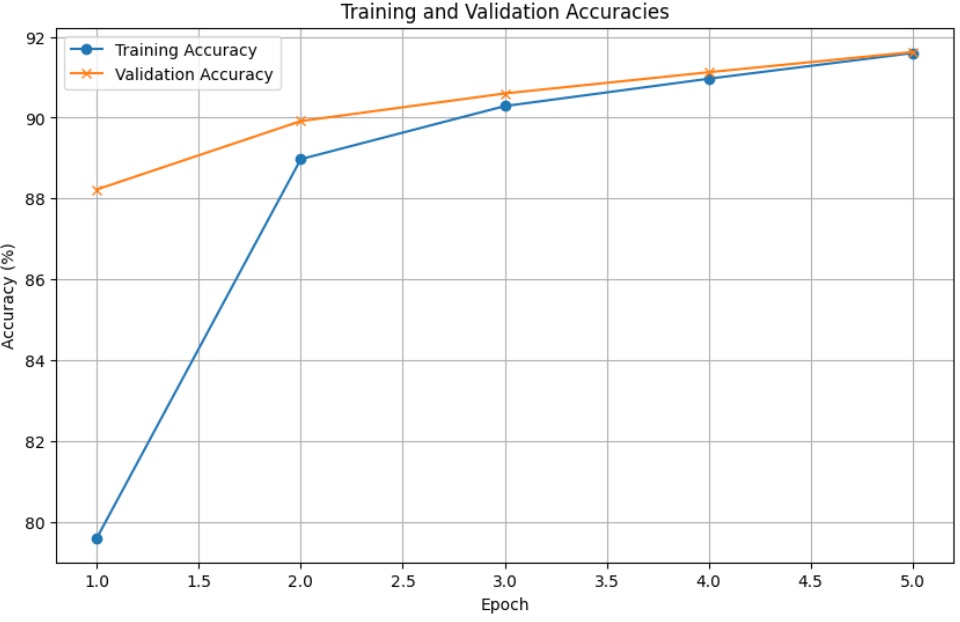
\
Logarithmic Number System Model Performance At Epoch 1: train-set accuracy at epoch 1: 52.00% Val-set accuracy at epoch 1: 24.00% At Epoch 2: train-set accuracy at epoch 2: 74.00% Val-set accuracy at epoch 2: 40.00% At Epoch 3: train-set accuracy at epoch 3: 86.00% Val-set accuracy at epoch 3: 58.00% At Epoch 4: train-set accuracy at epoch 4: 94.00% Val-set accuracy at epoch 4: 70.00% At Epoch 5: train-set accuracy at epoch 5: 96.00% Val-set accuracy at epoch 5: 68.00% elapsed time = 0.35 seconds.
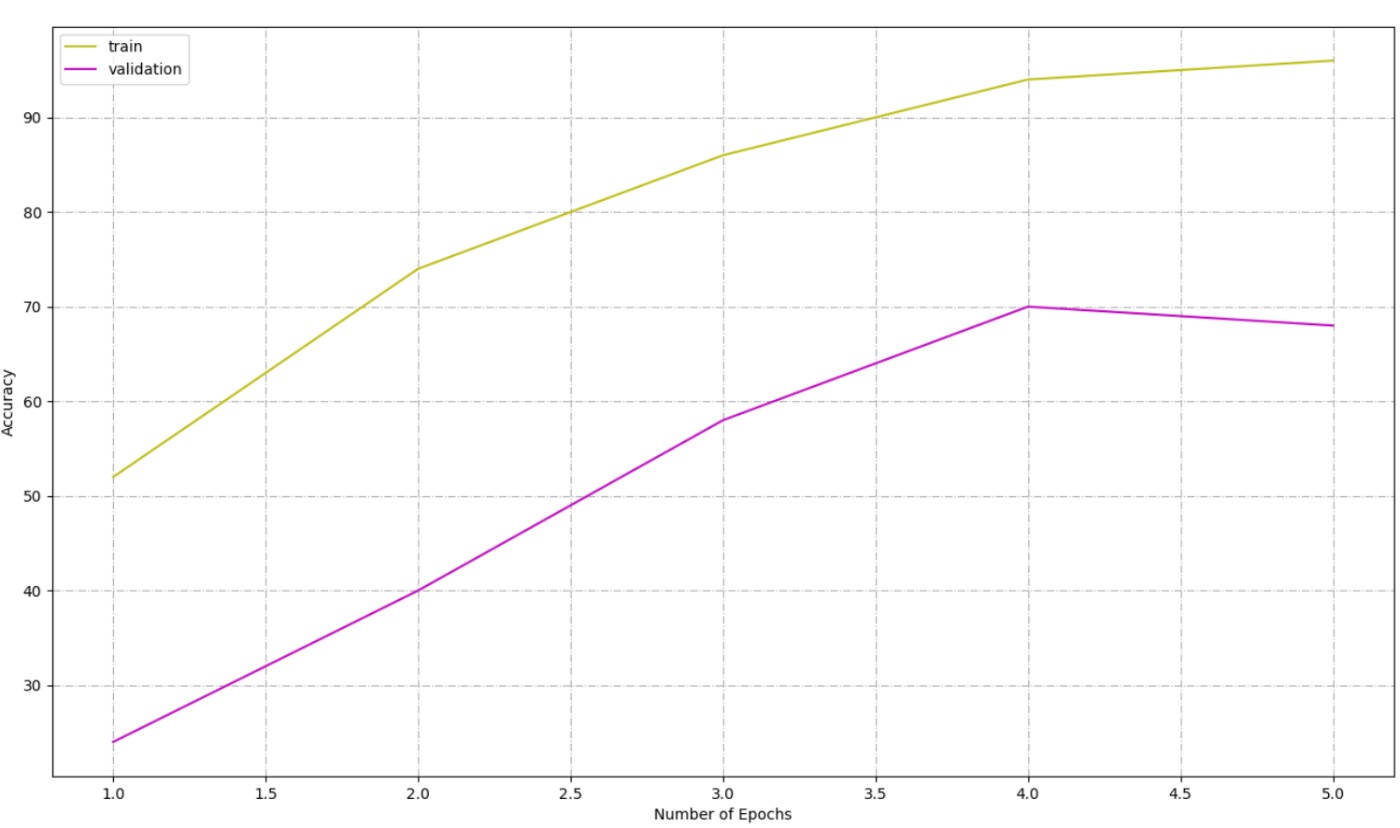
\
FP vs. LNS: Key Comparisons| Aspect | Floating-Point (FP) | Logarithmic Number System (LNS) | |----|----|----| | Training Time | 57.76s | 0.35s | | Train Accuracy | 91.60% | 96.00% | | Val Accuracy | 91.62% | 68.00% | | Precision | High | Lower (approximation errors) | | Memory Efficiency | Higher usage | Lower memory footprint | | Multiplication Handling | Native multiplication | Addition-based simplifications |
ConclusionThe tradeoffs between the Logarithmic Number System (LNS) and Floating-Point (FP) arithmetic present an interesting case study in hardware-software co-design for neural networks. While LNS offers significant advantages in certain areas:
Training Speed- Replaces multiplication with addition in the log domain
- Reduces complex operations to simpler arithmetic
- Particularly efficient for matrix multiplications in neural networks
- Can achieve 2–3x speedup in some implementations
- Typically requires fewer bits to represent numbers
- Can compress weights and activations more efficiently
- Reduces memory bandwidth requirements
- Lower power consumption for memory access
\ However, the accuracy challenges are significant:
- Precision loss during accumulation of small values
- Difficulty representing numbers very close to zero
- Potential instability in gradient calculations
- May require careful hyperparameter tuning
Several promising approaches could enhance the applicability of LNS:
1. Layer-Specific Arithmetic- Use FP for sensitive layers (like final classification)
- Apply LNS in computation-heavy hidden layers
- Dynamically switch based on numerical requirements
- Start training with FP for stability
- Gradually transition to LNS as weights converge
- Maintain critical paths in higher precision
- Custom accelerators with both FP and LNS units
- Smart scheduling between arithmetic types
- Specialized memory hierarchies for each format
- New activation functions optimized for LNS
- Modified optimization algorithms that maintain stability
- Hybrid number representations
To integrate LNS into deep learning frameworks, the following could be explored:
1. Custom Autograd Functions- Implement LNS operations as custom autograd functions
- Maintain gradient computation in the log domain
- Provide efficient CUDA kernels for acceleration
- Add native LNS tensor types
- Implement core operations (*+, -, , /) in the log domain
- Provide conversion utilities to/from floating point
- Create LNS versions of common layers (Linear, Conv2d)
- Optimize backward passes for LNS computation
- Support mixed precision training
\ The deep learning community could greatly benefit from integrating these capabilities into mainstream frameworks, enabling more efficient, low-power, and high-speed neural networks.
\ What are your thoughts on the balance between numerical precision and computational efficiency? Have you encountered specific use cases where LNS might be particularly beneficial?
\ Let me know your thoughts on this.
References\ [1] G. Alsuhli, et al., “Number Systems for Deep Neural Network Architectures: A Survey,” arXiv:2307.05035, 2023.
[2] M. Arnold, E. Chester, et al., “Training neural nets using only an approximate tableless LNS ALU.” 31st International Conference on Application-specific Systems, Architectures and Processors, IEEE, 2020, pp. 69–72. DOI
[3] O. Kosheleva, et al., “Logarithmic Number System Is Optimal for AI Computations: Theoretical Explanation of Empirical Success,” Paper
[4] D. Miyashita, et al., “Convolutional Neural Networks using Logarithmic Data Representation,” arXiv:1603.01025, Mar 2016.
[5] J. Zhao et al., “LNS-Madam: Low-Precision Training in Logarithmic Number System Using Multiplicative Weight Update,” IEEE Transactions on Computers, vol. 71, no. 12, pp. 3179–3190, Dec. 2022. DOI
- Home
- About Us
- Write For Us / Submit Content
- Advertising And Affiliates
- Feeds And Syndication
- Contact Us
- Login
- Privacy
All Rights Reserved. Copyright , Central Coast Communications, Inc.